Does my tree look good in this forest?
Judging trees by their appearance, knowing their family trees, and how and where they are growing, are essential elements for productive future forests.
Our phenotype is what we look like, whether tall, long-limbed or hairy. Phenotypes are an accurate and precise description of appearance, and are influenced by our genes and environment.
In plant breeding, understanding how appearance is affected by genes and environment can speed up genetic improvement programmes.
But measuring mature trees in a forest is more difficult than quantifying the appearance of an African violet in a greenhouse. Trees are big and difficult to measure accurately, there are operational limits in the number of trees and sites that can be measured, and separating genetic effect from environmental variation is not straightforward. These challenges of characterising individual trees is restricting current tree breeding.
Relieving the bottleneck
Recent technological advances in remote sensing, genomics and layers of geographic information system (GIS) data such as climate, soil type and forest management activities can be combined to relieve the phenotyping bottleneck.
Scion geneticists, remote sensing specialists and information technologists describe how they want to use these advances to develop a ‘phenotyping platform’ for forest trees in a new paper.
Why a phenotyping platform?
The overall aim of a phenotyping platform approach is to increase forest productivity matching trees with advanced genetics to the environment they will be growing in. In other words, the right tree in the right place.
Specifically, a phenotyping platform will allow monitoring of productivity changes that can be attributed to genetic improvement (realised genetic gain). It will also increase the number of trees breeders can select from as more trees are measured and extreme phenotypes are identified. This may be particularly useful for identifying disease resistance and responses to pest incursion.
The primary drivers of planted forestry productivity will become better understood as the scale and dimension of forest monitoring changes, with genotype productivity being able to be mapped across whole forests, not just at individual sample sites, and changes in growth, health and form monitored with time.
How might it work?
The research team has worked with Kaingaroa Forest as model to develop their concept and show its potential for maximising genetic gain in forests. Kaingaroa Forest in the central North Island is the largest plantation forest in New Zealand and planted mostly in radiata pine.
Focusing on areas of the forest where genetic information and forest management history were known, remote sensing was used to model phenotype traits such as tree height, and diameter. Productivity was calculated from canopy height derived from LiDAR data. Then layers of GIS data such as climate, terrain and stand management were added, and all the data was compiled into a form suitable for analysis.
Initial indications from the prototype phenotyping platform are that genetic improvement and climate had the strongest influence on the productivity of the trees in the study.
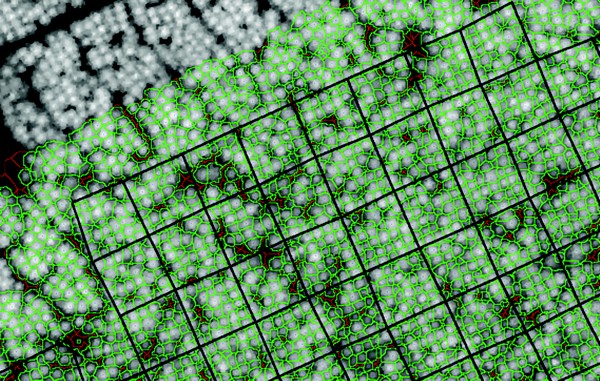
A greyscale image created from LiDAR delineating more than 2000 individual tree crowns, outlined in green, in Kaingaroa Forest.
Dungey, H. S., Dash, J. P., Pont, D., Clinton, P. W., Watt, M. S., & Telfer, E. J. (2018). Phenotyping Whole Forests Will Help to Track Genetic Performance. Trends in Plant Science. https://doi.org/10.1016/j.tplants.2018.08.005
